5. Ingestion service#
This section provides information regarding the ingestion service, one of the service component of BrainKB.
5.1. Overview#
Fig. 5.1 illustrates the architecture of the ingestion service, which follows the producer-consumer pattern and leverages RabbitMQ for scalable data ingestion. The service is composed of two main components: (i) the producer and (ii) the consumer. The producer component exposes API endpoints (see Fig. 5.2) that allow clients or users to ingest data. Currently, it supports the ingestion of KGs represented in JSON-LD and Turtle formats. Users can ingest raw JSON-LD data as well as upload files, either individually or in batches. At present, the ingestion of other file types, such as PDF, text, and JSON, has been disabled due to the incomplete implementation of the required functionalities. The consumer retrieves ingested data from RabbitMQ, processes it, and forwards it to the query service via API endpoints. The query service then inserts the processed data into the graph database.

Fig. 5.1 Architecture of the Ingestion Service.#
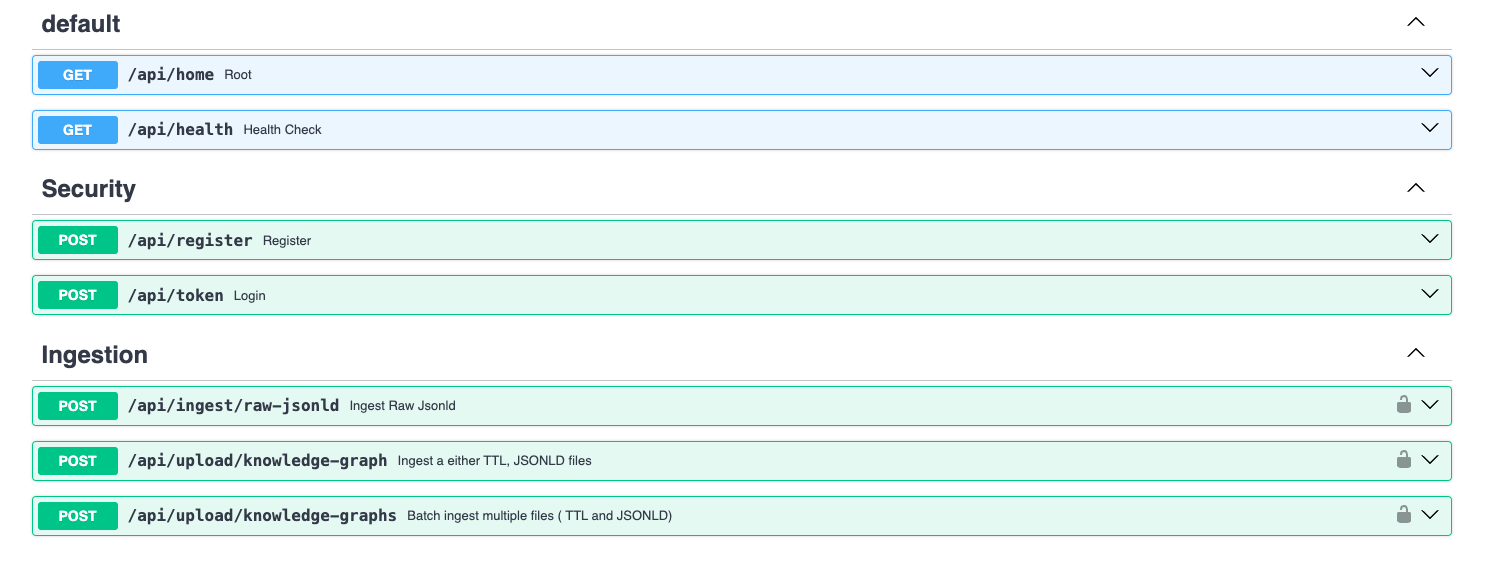
Fig. 5.2 Currently Enabled API Endpoints.#
5.2. Sequence Diagram#
This sequence diagram illustrates the data ingestion pipeline, detailing the process of how a client submits data, which is subsequently validated, processed, and stored in a graph database.
5.2.1. Producer Workflow Overview#
The diagram below highlights the producer in this pipeline, detailing each step in the process as described below.
sequenceDiagram
rect rgb(255,255,255)
box MistyRose Client
participant Client as Client/User
end
box Thistle Producer
participant API as Producer API
participant Validator as Shared
participant Publisher as RabbitMQ Publisher
end
box LightGoldenRodYellow RabbitMQ
participant RabbitMQ as RabbitMQ
end
%% Client submits data
Client->>API: 1. POST data (JSON-LD, TTL, etc.)
activate API
%% Producer service validates and publishes
API->>Validator: 2. Validate data format
activate Validator
Validator-->>API: Validation result
deactivate Validator
alt is valid data
API->>Publisher: 3. Format data for publishing
activate Publisher
Publisher->>RabbitMQ: 4. Publish message to queue
Publisher-->>API: Publish confirmation
deactivate Publisher
API-->>Client: 200 OK Response
else invalid data
API-->>Client: 400 Bad Request
end
deactivate API
end
5.2.1.1. Receiving Data from the Client#
The client initiates the ingestion process by submitting a
POSTrequest to the Producer API.The request contains structured data, typically in formats like JSON, JSON-LD, or TTL (Turtle).
Note: Support for additional formats, such as PDF and text, will be enabled once the necessary functionalities are fully developed (see Source codes of BrainKB Microservices) and integrated.
5.2.1.2. Validation & Preprocessing#
The Producer API passes the received data to the Shared (or shared.py that implements shared functionalities), which performs essential validation checks:
Ensuring the existence of the named graph in the database and ensuring the ingested data is in correct format, e.g., valid JSON-LD format.
It is important to note that to be able to proceed with ingestion, the client must either register a new named graph IRI (using the query service API endpoint) or select an existing one. This approach enables versioning, ensuring efficient data management and traceability.
If validation fails, the system returns a
400 Bad Requestto the client.
5.2.1.3. Publishing Data to RabbitMQ#
Once validated, the Producer RabbitMQ Publisher formats the data for ingestion.
The formatted data is published to RabbitMQ, which acts as a message broker to decouple producers and consumers.
A successful message publication triggers a publish confirmation, which is sent back to the Producer API.
5.2.2. Consumer Workflow Overview#
The diagram below highlights the consumer in this pipeline, detailing each step in the process as described below.
sequenceDiagram
rect rgb(255,255,255)
box LightGoldenRodYellow RabbitMQ
participant RabbitMQ as RabbitMQ Queue
end
box HoneyDew Consumer
participant Consumer as Listener
participant Processor as Shared
end
box AliceBlue Query Service
participant QueryService as Query Service
end
box Wheat Graph Database
participant GraphDB as Oxigraph
end
%% Worker service processes the message
RabbitMQ->>Consumer: 1. Consume message
activate Consumer
Consumer->>Processor: 2. Process data
activate Processor
%% Processing steps
Processor->>Processor: 3. Add provenance metadata
%% Acknowledge message
Processor-->>Consumer: Processing complete, provenance attached
%% Send to query service
Consumer->>QueryService: 4. Send processed data
activate QueryService
%% Store in database
QueryService->>GraphDB: 5. Store in graph database
activate GraphDB
GraphDB-->>QueryService: Storage confirmation
deactivate GraphDB
QueryService-->>Processor: Processing confirmation
deactivate QueryService
%% Acknowledge message
Processor-->>Consumer: Processing complete
deactivate Processor
Consumer-->>RabbitMQ: 6. Acknowledge message
deactivate Consumer
end
5.2.2.1. Message Consumption from RabbitMQ#
The RabbitMQ Queue holds messages published by the Producer.
The Consumer Listener picks up an available message from the RabbitMQ queue.
5.2.2.2. Adding Provenance Metadata#
The Consumer forwards the message to the Shared (or shared.py that implements shared functionalities) for further handling, e.g., adding the provenance information.
Example: Consider the following ingested data in TTL representation.
@prefix NCBIAssembly: <https://www.ncbi.nlm.nih.gov/assembly/> . @prefix NCBIGene: <http://identifiers.org/ncbigene/> . @prefix bican: <https://identifiers.org/brain-bican/vocab/> . @prefix biolink: <https://w3id.org/biolink/vocab/> . @prefix dcterms: <http://purl.org/dc/terms/> . @prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> . @prefix schema1: <http://schema.org/> . bican:000015fd3d6a449b47e75651210a6cc74fca918255232c8af9e46d077034c84d a bican:GeneAnnotation ; rdfs:label "LOC106504536" ; schema1:identifier "106504536" ; bican:molecular_type "protein_coding" ; bican:referenced_in bican:d5c45501b3b8e5d8b5b5ba0f4d72750d8548515c1b00c23473a03a213f15360a ; biolink:category bican:GeneAnnotation ; biolink:in_taxon bican:7d54dfcbd21418ea26d9bfd51015414b6ad1d3760d09672afc2e1e4e6c7da1dd ; biolink:in_taxon_label "Sus scrofa" ; biolink:symbol "LOC106504536" ; biolink:xref NCBIGene:106504536 .
A new property
prov:wasInformedBy, is added to the initial TTL data, establishing a link to the triple that contains provenance information, as illustrated below. Please note that the provenance is attached for all the triples. For example, if you are uploading a TTL file that contains 30 triples then all the 30 triples will have provenance information attached.<https://identifiers.org/brain-bican/vocab/ingestionActivity/e4db1e0b-98ff-497c-88b1-afb4a6d7ee14> a prov:Activity, bican:IngestionActivity ; prov:generatedAtTime "2025-01-31T16:52:22.061674+00:00"^^xsd:dateTime ; prov:wasAssociatedWith bican:000015fd3d6a449b47e75651210a6cc74fca918255232c8af9e46d077034c84d, bican:00027255beed5c235eaedf534ac72ffc67ed597821a5b5c0f35709d5eb93bd47, <https://identifiers.org/brain-bican/vocab/agent/testuser> . <https://identifiers.org/brain-bican/vocab/provenance/e4db1e0b-98ff-497c-88b1-afb4a6d7ee14> a prov:Entity ; dcterms:provenance "Data posted by testuser on 2025-01-31T16:52:22.061674Z" ; prov:generatedAtTime "2025-01-31T16:52:22.061674+00:00"^^xsd:dateTime ; prov:wasAttributedTo <https://identifiers.org/brain-bican/vocab/agent/testuser> ; prov:wasGeneratedBy <https://identifiers.org/brain-bican/vocab/ingestionActivity/e4db1e0b-98ff-497c-88b1-afb4a6d7ee14> .
Final data after adding provenance information.
@prefix NCBIGene: <http://identifiers.org/ncbigene/> . @prefix bican: <https://identifiers.org/brain-bican/vocab/> . @prefix biolink: <https://w3id.org/biolink/vocab/> . @prefix dcterms: <http://purl.org/dc/terms/> . @prefix prov: <http://www.w3.org/ns/prov#> . @prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> . @prefix schema1: <http://schema.org/> . @prefix xsd: <http://www.w3.org/2001/XMLSchema#> . bican:000015fd3d6a449b47e75651210a6cc74fca918255232c8af9e46d077034c84d a bican:GeneAnnotation ; rdfs:label "LOC106504536" ; schema1:identifier "106504536" ; prov:wasInformedBy <https://identifiers.org/brain-bican/vocab/provenance/e4db1e0b-98ff-497c-88b1-afb4a6d7ee14> ; #this links to the new provenance information bican:molecular_type "protein_coding" ; bican:referenced_in bican:d5c45501b3b8e5d8b5b5ba0f4d72750d8548515c1b00c23473a03a213f15360a ; biolink:category bican:GeneAnnotation ; biolink:in_taxon bican:7d54dfcbd21418ea26d9bfd51015414b6ad1d3760d09672afc2e1e4e6c7da1dd ; biolink:in_taxon_label "Sus scrofa" ; biolink:symbol "LOC106504536" ; biolink:xref NCBIGene:106504536 . #added new provenance information regarding the ingestion activity. <https://identifiers.org/brain-bican/vocab/ingestionActivity/e4db1e0b-98ff-497c-88b1-afb4a6d7ee14> a prov:Activity, bican:IngestionActivity ; prov:generatedAtTime "2025-01-31T16:52:22.061674+00:00"^^xsd:dateTime ; prov:wasAssociatedWith bican:000015fd3d6a449b47e75651210a6cc74fca918255232c8af9e46d077034c84d, bican:00027255beed5c235eaedf534ac72ffc67ed597821a5b5c0f35709d5eb93bd47, <https://identifiers.org/brain-bican/vocab/agent/testuser> . <https://identifiers.org/brain-bican/vocab/provenance/e4db1e0b-98ff-497c-88b1-afb4a6d7ee14> a prov:Entity ; dcterms:provenance "Data posted by testuser on 2025-01-31T16:52:22.061674Z" ; prov:generatedAtTime "2025-01-31T16:52:22.061674+00:00"^^xsd:dateTime ; prov:wasAttributedTo <https://identifiers.org/brain-bican/vocab/agent/testuser> ; prov:wasGeneratedBy <https://identifiers.org/brain-bican/vocab/ingestionActivity/e4db1e0b-98ff-497c-88b1-afb4a6d7ee14> .
5.2.2.3. Sending Processed Data to the Query Service#
The Consumer forwards the processed data to the Query Service.
The Query Service acts as an interface to the Graph Database, enabling operations such as querying and inserting data.
5.2.2.4. Storing Data in the Graph Database#
The Query Service sends the structured data to the Graph Database, which in our case is the Oxigraph.
The Graph Database confirms successful/unsuccessful storage operation.
5.2.2.5. Acknowledging Message Processing#
The Query Service sends a confirmation back to the Processor.
The Processor notifies the Consumer that processing is complete.
Finally, the Consumer acknowledges the message to RabbitMQ, ensuring:
The message is marked as processed.